Action 2 – Genetic characterisation of autochthonous chicken and turkey breeds farmed in Italy
Genetic characterisation of Italian chicken breeds with SNP markers will include the Italian Collo Nudo and Millefiori Piemontese breeds. Characterisation with SNP markers will also involve Italian breeds of guinea-fowl (Camosciata), duck (Mignon and Germanata Veneta), and goose (Padovana) (UniPD). The breeds considered along the two projects and the Partners involved are shown in the Table below.
Polymorphisms closely related to heat stress will be characterised, in order to highlight variability between chicken breeds (UniPG). Genetic diversity and population specific mutations will be investigated in turkeys through the acquisition of genome sequencing data (UniMI).
Moreover, the characterisation with molecular microsatellite markers of breeding birds reared at the Partners’ facilities will allow to identify candidate male breeders, to monitor variability within breeds, to identify new family lines, to complete the genetic characterisation of additional chicken breeds, and to evaluate the polymorphism of PAX7 gene, which is a candidate marker for some productive traits (consulting provided by UniTO).
To see the results from the first TuBAvI project, click here.
Autochthonous chicken, turkey, Guinea fowl, duck, and goose breeds undergoing genetic characterisation
| UniMI | UniFI | UniPD | UniPG | UniPI | UniTO | |
| Chicken breeds | Mericanel della Brianza | Mugellese Valdarnese bianca |
Ermellinata di Rovigo Robusta Lionata Robusta Maculata Millefiori di Lonigo Padovana Pepoi Polverara |
Ancona Livorno |
Livorno, Siciliana | Bianca di Saluzzo Bionda Piemontese |
| Turkey breeds | Brianzolo Bronzato comune Colli Euganei Ermellinato di Rovigo Nero d’Italia Parma e Piacenza Romagnolo |
|||||
| Guinea fowl breeds | Camosciata | |||||
| Duck breeds | Mignon, Germanata veneta | |||||
| Goose breeds | Padovana |
Action 2 – TuBAvI (2017-2020): results
Genetic characterisation with SNP markers: genetic analysis of chicken breeds (UniPD)
Breed consistency is evaluated by means of genetic diversity indexes, which are based on the presence of SNPs (Single-Nucletoide Polymorphisms) molecular markers in the genome. SNPs are a mutation of genetic material at the level of a single nucleotide, so that the polymorphic allele occurs in more than 1% of the population. Italian breeds show different degrees of genetic diversity, according to the different estimated indexes. The analysis of the results allows to provide information on the genetic variability within the breed and on the conservation status of the population.
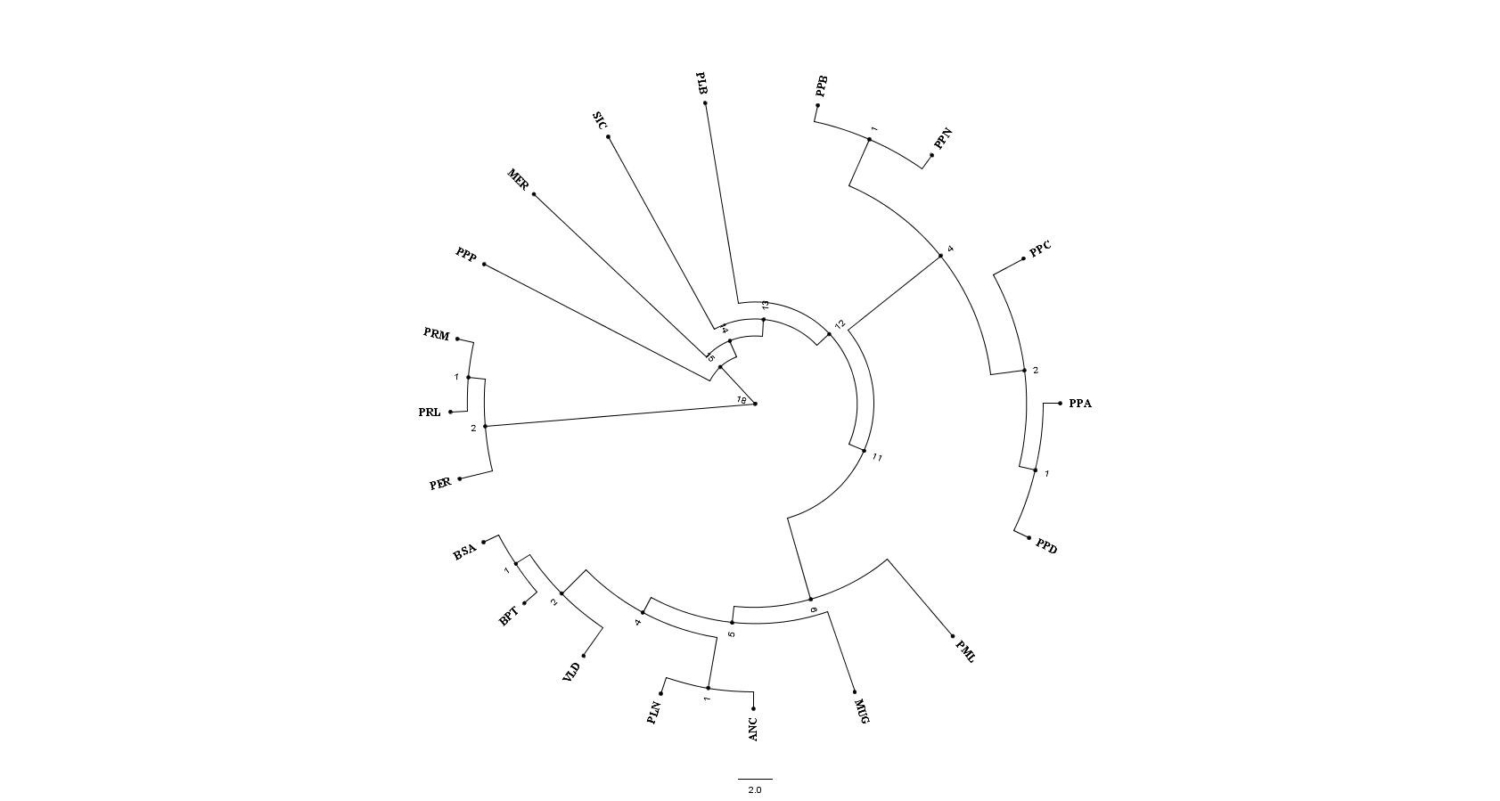
In addition, population studies performed by SNP molecular markers allow to estimate the genetic distances among breeds and work out a phylogenetic tree, corresponding to a diagram showing the lineage relationships among groups of species or breeds within species (Figure 1) (UniPD).
Every knot (branching) shows the most recent common point for the breed, that is the most recent ancestor of the subjects in the following knots. The length of the branches is strictly linked to evolutionary time, genetic diversity and genetic mutations occurring between them.
Genetic diversity indexes
MAF (minor allelic frequency): it ranges from 0 to 1 and identifies the fixation parameter of traits within a population (it tends to 0 for fixed traits)
He (expected heterozygosis): it shows the probability that a random subject within a population, that is in accordance with the Hardy-Weinberg equilibrium, is heterozygous for given traits; Ho (observed heterozygosis): it shows the real heterozygosis rate within the population for each allelic locus taken into consideration. Ho and He allow to analyze variability within the breed.
FHOM (inbreeding coefficient based on excess of homozygosis): it shows the fraction of individuals within a population with homozygosis variant. The more it tends to 1, the higher is the homozygosis.
ROH (Runs of homozygosity): specific regions of the genome with uninterrupted series of SNP markers with homozygous genotype. These regions of “continuous” homozygosis are inherited by parents (by mendelian inheritance) with a higher probability if they are relative to each other, so their dimensions and frequencies are higher in case of high inbreeding.
FROH (inbreeding coefficient based on ROH presence): inbreeding coefficient estimated on the basis of the presence of continuous homozygosity regions in the genome (ROH, Runs of homozygosity). FROH shows the proportion existing between total length of ROH in an individual genome and total length of the genome covered by the SNPs used for genetic characterisation.
Table 1 – Average genetic diversity indexes for the evaluation of breed consistency in rural breeding. The number in parentheses is the number of animals tested per breed. (UniPD)
| Indici | ANC (24) |
BSA (24) |
BPT (22) |
PER (23) |
PLB (24) |
PLN (24) |
MER (23) |
PML (23) |
MUG (24) |
PPA (24) |
PPC (24) |
PPD (24) |
PPP (24) |
PPB (24) |
PPN (24) |
PRL (23) |
PRM (24) |
SIC (24) |
VLD (24) |
| MAF | 0.267 | 0.286 | 0.283 | 0.309 | 0.269 | 0.263 | 0.282 | 0.281 | 0.284 | 0.241 | 0.238 | 0.247 | 0.277 | 0.260 | 0.257 | 0.305 | 0.304 | 0.259 | 0.283 |
| He | 0.274 | 0.336 | 0.317 | 0.220 | 0.218 | 0.231 | 0.261 | 0.291 | 0.300 | 0.146 | 0.179 | 0.232 | 0.168 | 0.248 | 0.213 | 0.185 | 0.166 | 0.123 | 0.322 |
| Ho | 0.263 | 0.339 | 0.325 | 0.199 | 0.205 | 0.233 | 0.232 | 0.293 | 0.281 | 0.151 | 0.169 | 0.219 | 0.154 | 0.216 | 0.201 | 0.181 | 0.157 | 0.129 | 0.321 |
| FHOM | 0.284 | 0.076 | 0.116 | 0.459 | 0.465 | 0.365 | 0.368 | 0.166 | 0.225 | 0.588 | 0.538 | 0.404 | 0.579 | 0.411 | 0.454 | 0.508 | 0.572 | 0.648 | 0.127 |
| FROH | 0.201 | 0.081 | 0.081 | 0.305 | 0.427 | 0.296 | 0.326 | 0.202 | 0.236 | 0.509 | 0.410 | 0.230 | 0.482 | 0.310 | 0.353 | 0.353 | 0.410 | 0.607 | 0.121 |
ANC Ancona, BSA Bianca di Saluzzo, BPT Bionda Piemontese, PER Ermellinata di Rovigo, PLB Livorno Bianca, PLN Livorno Nera, MER Mericanel della Brianza, PML Millefiori di Lonigo, MUG Mugellese, PPA Padovana Argentata, PPC Padovana Camosciata, PPD Padovana Dorata, PPP Pepoi, PPB Polverara Bianca, PPN Polverara Nera, PRL Robusta Lionata, PRM Robusta Maculata, SIC Siciliana, VLD Valdarnese
Genetic characterisation with SNP markers: genetic analysis of turkey breeds (UniMI)
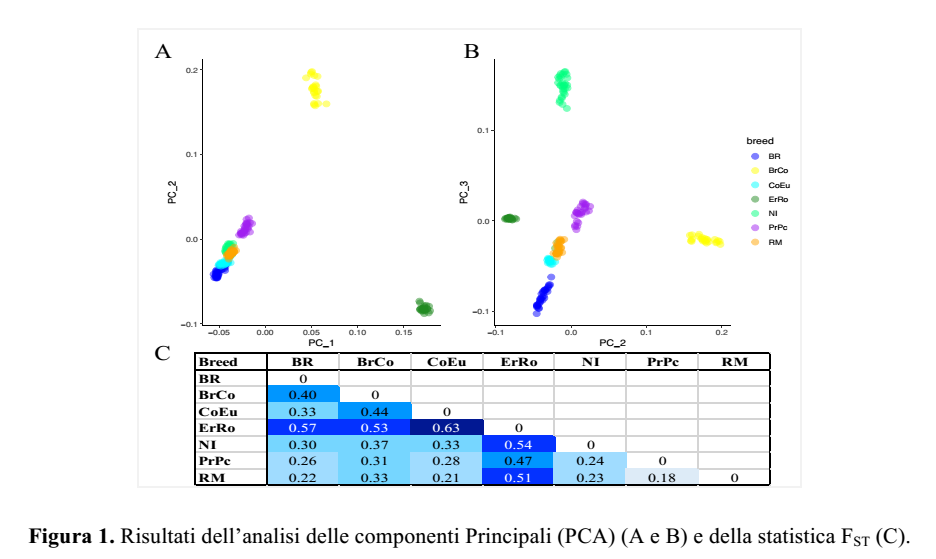
UniMI performed the genetic characterisation with high dense SNPs chip of seven Italian turkey breeds (Table 1 shows the number of birds in the samples). Results highlighted variability and genetic relationships among the breeds. In particular, Principal Component Analysis (Figure 1A PCA_1 vs PCA_2; Figure 1B PCA_2 vs PCA_3) clearly shows that individuals gather in clusters corresponding to their breeds. Ermellinato di Rovigo (ErRO) and Bronzato Comune (BrCO) are the breeds most separated from the others. This separation means that a marked genomic difference exists both between them and among the other breeds, which is confirmed also by FST values, shown in Figure 1C.
Parma e Piacenza (PrPc) and Romagnolo (RM) are the breeds with the lowest differentiation level (FST=0.18).
The genomic inbreeding indexes, FHOM and FROH, were calculated for all the birds in the different breeds. Both indexes showed variability both within and between the populations, suggesting high inbreeding values (average values per breed are shown in Table 2) for ErRo and CoEu breeds, and low inbreeding value for PrPc breed.
Statistic analysis: glossary
PCA (Principal Component Analysis): the Principal Component Analysis (PCA) is a statistical analysis procedure that allows to describe the variability of a sample through a small number of highly informative variables (the components). When applied to genomics, PCA allows to identify breed clusters specific for the mutations identified in the genome of the breeds (the variables).
FST: it estimates the degree of differentiation among populations on the basis of allelic frequences among subpopulations (comparison between groups). FST ranges from 0 to 1 (FST=1 highest differentiation; FST=0 no differentiation).